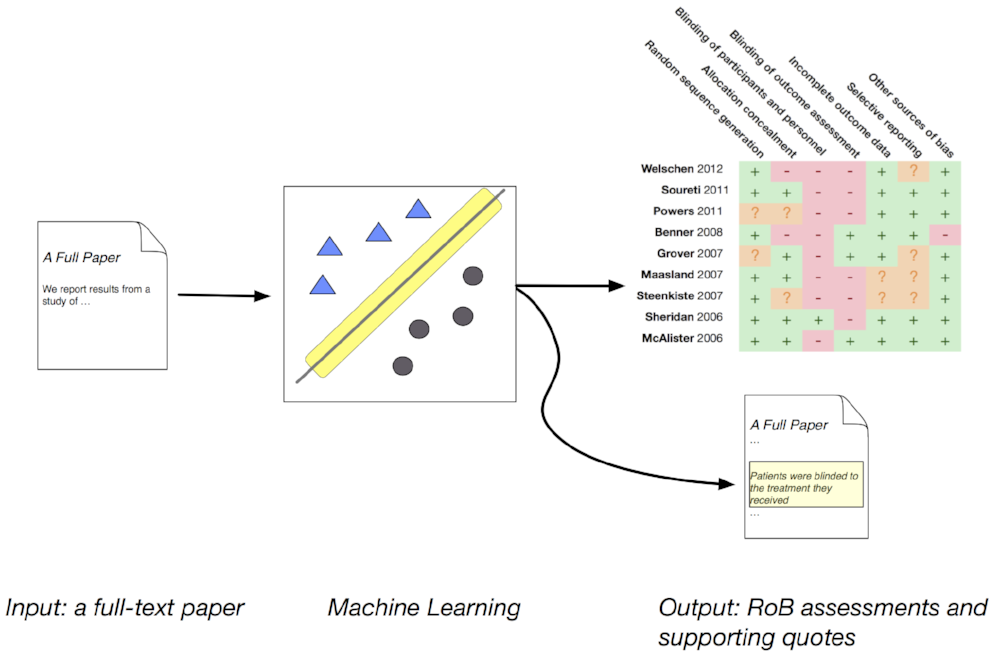
We are excited to unveil Trialstreamer, a living (continuously updated) database of all published randomized controlled trial reports, automatically annotated with data we extract via the machine learning models in RobotReviewer. To do this, we monitor PubMed and other sources daily for new literature, and use our previously validated machine learning model to automatically identify the subset of articles that constitute new RCT reports in humans.
Next we automatically extract from all identified trials: Sample sizes; Snippets of text that describe trial Populations, Interventions, Comparators and Outcomes (PICO elements), and normalized terms for these; An estimated proxy for study quality, as a (simplified) overall “risk of bias” score; And the “punchlines” that seem to convey the main trial findings, along with an inferred directionality of said finding.
We make this database (updated daily) available directly. As of this writing, it comprises 697,217 trials. We have also implemented a simple faceted search interface that facilitates browsing the aggregated evidence. Try it out here: https://trialstreamer.robotreviewer.net/.
This is just one potential use of the data, however. The resource might also permit novel views of the evidence base that it might afford. For example, in an ACL 2020 Demo paper led by Ben Nye, we used the underlying data to automatically construct “evidence maps” for queries, on-demand:
Elsewhere, Trialstreamer data has been incorporated into the neural covidex search engine for Covid to complement the body of non-RCT literature pertaining to COVID-19.
Aside from more efficient navigation of the evidence base, we are optimistic that the semi-structured data automatically extracted from the underlying trial reports might afford novel analyses of the trials literature. For example, this readily allows for an analysis of the interventions studied over time, or of the (estimated) risks of bias of trials in particular sub-areas. We’d be keen to hear other potential uses and extensions; so please reach out to us if you have questions or thoughts.
More details are available in the JAMIA paper describing Trialstreamer, here: https://academic.oup.com/jamia/advance-article/doi/10.1093/jamia/ocaa163/5907063.